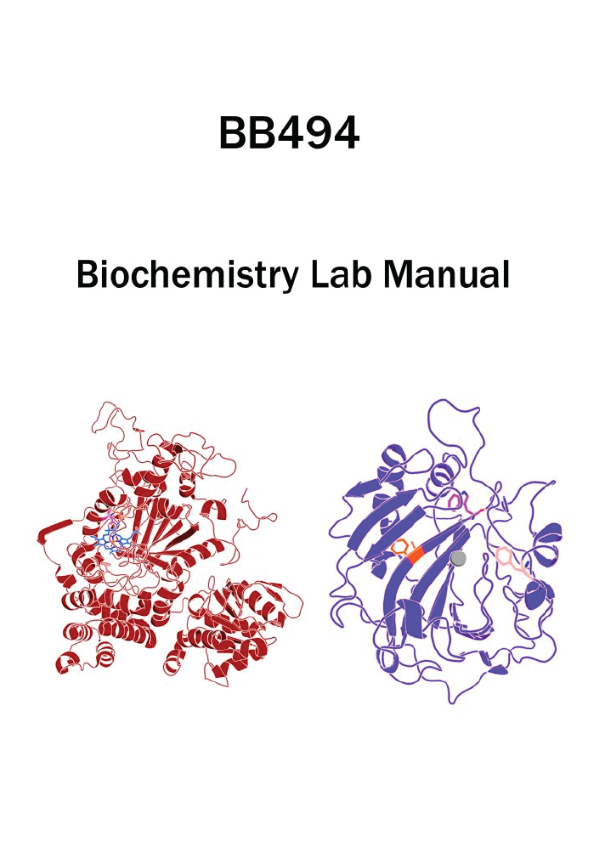
Your goal is to design a meaningful scientific study surrounding one of the enzymes that has been chosen for you: thermostable human carbonic anhydrase II or E. coli catalase HPII. Traditionally, biochemists are able to change any amino acid in a protein to any of the other naturally occurring amino acids by standard mutagenesis methods. This is common to facilitate the understanding of how a protein works (regulation, binding etc.) and to improve its utility for other applications (stability, activity etc.). Here, we will break from tradition and enable you to select from new amino acids structures (non-canonical amino acids) to study proteins in ways that were not possible before. One could put the ncAAs anywhere in the protein via genetic code expansion, but because of time constrains of mutagenesis, we are forced to select some sites for you to explore. The key to this endeavor is to identify what is important about the sites we have selected for you regarding the proteins’ stability, activity, regulation, etc. and then select from the new chemical ability of the ncAAs to develop a meaningful scientific study that has never been possible before.
Using the list of available mutation sites and ncAAs provided by the instructor (Appendix 6), choose one or two sites to explore by the incorporation of ncAAs. You will be able to generate two new mutant proteins to study in parallel with the wild-type protein. Searching the literature on your particular enzyme can highlight particular residues in that enzyme that are thought to have implications on the protein’s structure or function. Carefully consider the chemical properties (i.e., the wild-type amino acid side chain) that already exist at the site(s), and the new properties that may be introduced by the ncAA. Is the site supposedly involved in the catalysis of the enzyme? Is it responsible for maintaining structural characteristics of the protein, or involved in relaying structural changes to different areas of the protein? These are important aspects to consider when selecting the sites and ncAAs that will be studied for the entire term. Be sure to base your study in the context of the scientific literature.
To facilitate the production and purification of the chosen ncAA-mutant proteins, the genes of interest a thermostable variant of human carbonic anhydrase II (CA) as well as E. coli catalase HPII (HPII) were commercially synthesized to optimize their codon usage for expression in E. coli. Both genes were cloned into the pBad expression plasmid. The cloning event removes the stop codon and adds a C-terminal histidine-rich affinity tag, allowing easy purification of the protein product. A stop codon (TAG) was then incorporated in place of a codon in the wild-type gene by using mutagenic primers. These two plasmids—one containing the wild-type gene, and the other with the TAG mutation—will allow production of wild-type and ncAA-mutant versions of the protein, respectively. With the set of pure proteins (wild-type and ncAA-mutant), each group will perform comparison studies of the structure and function of the wild-type and ncAA-mutant proteins. Relevant genetic sequences for each protein can be found in the corresponding tables in Appendix 2, and available ncAA structures can be found in Appendix 3. Using these genetic sequences, the predicted molecular weight and protein product sequence can be determined using the provided web resource. This information will be useful in later steps of the purification process.